
|
Working in the lab. From left to right: Dr. Brauns, Aaron Stancik, and Dave Church |
RNA folding
RNA plays an essential role in numerous biochemical processes and is much more than an academic curiosity. Recent evidence demonstrates that RNA is not a passive participant in the regulation of gene expression as previously thought. Untranslated regions of some mRNA transcripts can adopt an alternative conformation in the presence of a specific substrate. These have been termed “riboswitches,” and the conformational change regulates the expression of the associated gene by turning its translation either on or off. Expansions of trinucleotide repeats (e.g., [CUG]n, [GAA]n, etc.) found in some mRNA transcripts have been linked to diseases ranging from fragile X syndrome to Huntington’s disease. While their pathological role remains unclear, they tend to form stable secondary structures that are likely related to functionality. In addition, there are RNA viruses (retro viruses); two well known examples include HIV and hepatitis B. On a slightly different note, it has been postulated that the earliest forms of life on earth were RNA based organisms. If true, then studying RNA folding will open a window through which to observe some of the most fundamental aspects of biochemistry.
The biological relevance of RNA and its potential role as a therapeutic target underscore the need to systematically understand the rules that dictate RNA structure and dynamics. The origin of the “RNA folding problem” has traditionally been rooted in structure prediction. However, equilibrium structure prediction by itself is insufficient; the RNA folding problem reaches much farther. RNA in its biological context is neither static nor rigid. It is in a constant state of flux, interconverting between closely related conformations in response to molecular demands. The dynamic character adds to its structural intricacy and augments its functionality. Hence, genuinely useful knowledge of a structure involves knowing where the atoms are, how much energy it takes to move them, how fast they move, and what factors influence their motion (i.e., knowledge of its folding landscape). We need to know how the structures form, not just that they form. If we have any hope of manipulating an RNA structure to alter its function (e.g., for therapeutic purposes) or to deduce function based on structure, we need to thoroughly explore the energy landscape of RNA. Doing so will advance our understanding of RNA more than equilibrium structural studies alone.
Time-resolved infrared spectroscopy
We use a laser induced temperature jump (T-jump) to perturb the thermal equilibrium between the folded and unfolded ensembles of an RNA sample. The subsequent conformational changes are monitored using time resolved infrared (IR) spectroscopy. The instrumentation in my laboratory permits us to observe transient IR signals with ~10 ns resolution. Using IR spectroscopy is advantageous since the spectral changes are monitored at different wavenumbers that correspond to certain vibrational transitions of the RNA.
Since a laser induced T-jump instrument is not commercially available. We use a combination of commercial components and custom made components to build the instrumentation in our lab. The figure below is a schematic representation of the experimental apparatus.
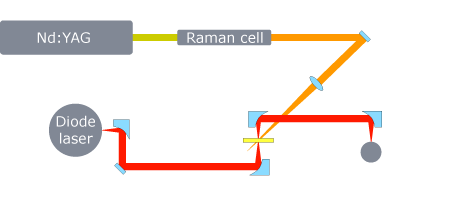 Essentially, it is a pump-probe experiment. The pump is a < 10 ns (FWHM) pulse from a Q-switched Nd:YAG laser operating at 10 Hz. The YAG fundamental (1.064 µm) is Raman shifted in H2 to a wavelength of 1.9 µm. The solvent (typically D2O) absorbs ~20% of the pump pulse energy and deposits it as heat (RNA does not absorb at this wavelength). A ~20 mJ pulse can raise the temperature of the laser interaction volume by as much as 25°C. The thermal equilibration time of the D2O is about 50 ps. Hence, a 10 ns pulse results in a virtually instantaneous temperature increase that displaces the thermal equilibrium between the folded and unfolded ensembles in favor of the unfolded state. The temperature remains constant out to about 1 ms, ultimately limiting the long time response of the system. Thin optical path lengths (< 100 µm) ensure uniform heating. The pump passes through a polarizer that can be rotated to finely adjust the magnitude of the T-jump. The probe beam is focused tightly on the sample and intersects the pump. The diameter of the pump beam on the sample is adjusted to be ~5× the diameter of the probe beam using a long focal length lens (this minimizes fluctuations due to any beam drift). A CW (continuous wave) lead-salt diode laser tuned to specific vibrational transitions of the RNA is used to probe the ensuing relaxation dynamics. The transmitted signal is collected and focused onto a mercury cadmium telluride (MCT) detector. The detector has a 100 MHz bandwidth resulting in a ~10 ns rise time (a good match for the pump pulse width). The detector signal is digitized by a 12-bit, high-speed data acquisition card. Gold coated, off-axis paraboloidal mirrors are used throughout the probe beam and signal paths. A LabVIEW program is used to collect the data and coordinate the instrumentation.
Essentially, it is a pump-probe experiment. The pump is a < 10 ns (FWHM) pulse from a Q-switched Nd:YAG laser operating at 10 Hz. The YAG fundamental (1.064 µm) is Raman shifted in H2 to a wavelength of 1.9 µm. The solvent (typically D2O) absorbs ~20% of the pump pulse energy and deposits it as heat (RNA does not absorb at this wavelength). A ~20 mJ pulse can raise the temperature of the laser interaction volume by as much as 25°C. The thermal equilibration time of the D2O is about 50 ps. Hence, a 10 ns pulse results in a virtually instantaneous temperature increase that displaces the thermal equilibrium between the folded and unfolded ensembles in favor of the unfolded state. The temperature remains constant out to about 1 ms, ultimately limiting the long time response of the system. Thin optical path lengths (< 100 µm) ensure uniform heating. The pump passes through a polarizer that can be rotated to finely adjust the magnitude of the T-jump. The probe beam is focused tightly on the sample and intersects the pump. The diameter of the pump beam on the sample is adjusted to be ~5× the diameter of the probe beam using a long focal length lens (this minimizes fluctuations due to any beam drift). A CW (continuous wave) lead-salt diode laser tuned to specific vibrational transitions of the RNA is used to probe the ensuing relaxation dynamics. The transmitted signal is collected and focused onto a mercury cadmium telluride (MCT) detector. The detector has a 100 MHz bandwidth resulting in a ~10 ns rise time (a good match for the pump pulse width). The detector signal is digitized by a 12-bit, high-speed data acquisition card. Gold coated, off-axis paraboloidal mirrors are used throughout the probe beam and signal paths. A LabVIEW program is used to collect the data and coordinate the instrumentation.
|
