Barrie D. Robison
Professor of Biological Sciences
 Polymorphic Games uses the principles of biological evolution to develop superior video games. Traditional video games are usually scripted, featuring “waves” of enemies that have defined and predictable characteristics. A player’s success in such games is based on learning the predictable, rote script necessary to advance to subsequent levels. By integrating principles of evolutionary biology, video games can be made more compelling. For example, our current prototypes feature generations of enemies that undergo adaptation through natural selection. The enemies with the traits that best counter the player’s strategies survive to reproduce, and their offspring feature prominently in the next generation (analogous to a game level or wave). We essentially create an evolutionary arms race between the player and the enemy population. The success of the player is based on her comprehension and application of principles of evolutionary biology. The parallels to real world examples are numerous, and include the rapid evolution of antibiotic resistance in microbial pathogens, adaptation of crop pests to chemical and biological control measures, and behavioral adaptation to captivity in domesticated animals.
Polymorphic Games uses the principles of biological evolution to develop superior video games. Traditional video games are usually scripted, featuring “waves” of enemies that have defined and predictable characteristics. A player’s success in such games is based on learning the predictable, rote script necessary to advance to subsequent levels. By integrating principles of evolutionary biology, video games can be made more compelling. For example, our current prototypes feature generations of enemies that undergo adaptation through natural selection. The enemies with the traits that best counter the player’s strategies survive to reproduce, and their offspring feature prominently in the next generation (analogous to a game level or wave). We essentially create an evolutionary arms race between the player and the enemy population. The success of the player is based on her comprehension and application of principles of evolutionary biology. The parallels to real world examples are numerous, and include the rapid evolution of antibiotic resistance in microbial pathogens, adaptation of crop pests to chemical and biological control measures, and behavioral adaptation to captivity in domesticated animals.
Adding biological evolution to video games makes the games better for the game player and facilitates player comprehension of complex concepts that are hard to teach.
The past ten years have seen an explosion of studies seeking to unravel the molecular details of adaptive evolution. Although these studies provide insight into the genetic basis of many interesting traits, they have done little to help us better understand important evolutionary processes or predict molecular and phenotypic evolution. In fact, the usual approach in genomic studies of adaptation, in which we attempt to find biological meaning among the lists of genes underlying behavioral evolution, may be inherently backward.
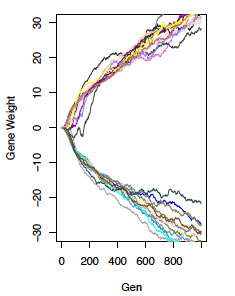
We are using simulation models of artificial neural networks to make predictions regarding the genetic manifestation of selection on behavioral phenotypes. We hope to use these results to provide an interpretive framework for genomic studies of behavioral adaptation. We are particularly interested in questions regarding the repeatability and predictability of evolution. Such questions include:
1) given similar selection pressures, how often will two taxa evolve the same molecular route to the new phenotypic optimum?
2) To what extent does this repeatability depend on topology of the underlying neural networks?
3) How often should we observe genes of large effect, versus diffuse and highly polygenic inheritance, and do these outcomes depend on underlying network features?
 The process of adaptation to captivity can cause striking changes in a variety of behavioral phenotypes, including aggressive, feeding, and reproductive behaviors. However, some of the most profound and consistent evolutionary changes occur in behaviors related to fearfulness and anxiety, with domesticated animals often described as less fearful than their wild progenitor populations. The evolution of reduced fearfulness during captivity has been observed in a variety of vertebrate taxa, including mammals (Harri et al., 2003), birds (Jensen & Andersson, 2005), and fish (Johnsson & Abrahams, 1991; Robison & Rowland, 2005). Despite the prevalence of behavioral evolution during adaptation to captivity, we know surprisingly little about the underlying genetic changes that occur during domestication. What kinds of molecular variation (amino acid sequence, regulatory, or non coding RNA) are associated with variation in fear and anxiety related behaviors? What kinds of genes are typically changed in response to domestication selection? Are the apparently parallel changes in behavior across independent domestication events reflective of parallel molecular evolution, or does domestication result from myriad molecular routes to the same phenotypic endpoint? The answers to these questions are important in many contexts, including the study of the molecular mechanisms of behavioral evolution, the genetic basis of complex behaviors, and the effective conservation of captively reared species.
The process of adaptation to captivity can cause striking changes in a variety of behavioral phenotypes, including aggressive, feeding, and reproductive behaviors. However, some of the most profound and consistent evolutionary changes occur in behaviors related to fearfulness and anxiety, with domesticated animals often described as less fearful than their wild progenitor populations. The evolution of reduced fearfulness during captivity has been observed in a variety of vertebrate taxa, including mammals (Harri et al., 2003), birds (Jensen & Andersson, 2005), and fish (Johnsson & Abrahams, 1991; Robison & Rowland, 2005). Despite the prevalence of behavioral evolution during adaptation to captivity, we know surprisingly little about the underlying genetic changes that occur during domestication. What kinds of molecular variation (amino acid sequence, regulatory, or non coding RNA) are associated with variation in fear and anxiety related behaviors? What kinds of genes are typically changed in response to domestication selection? Are the apparently parallel changes in behavior across independent domestication events reflective of parallel molecular evolution, or does domestication result from myriad molecular routes to the same phenotypic endpoint? The answers to these questions are important in many contexts, including the study of the molecular mechanisms of behavioral evolution, the genetic basis of complex behaviors, and the effective conservation of captively reared species.
We use the zebrafish to study the genetic basis of fear related behaviors that commonly change during domestication in fish. The zebrafish is an excellent system for this purpose, combining a robust genomics toolset with superior experimental tractability. Our lab has documented variation among wild and laboratory derived strains consistent with the effects of domestication. The behavioral variation among wild and domesticated strains can be seen in the video on this page. This video shows two tanks of zebrafish randomly pulled from our zebrafish colony. One tank contains a highly domesticated (>30 generations) strain (Scientific Hatcheries), and the other tank contains a strain with a more recent history (~5 generations) of domestication (Nadia). We have quantified surface orientation (Robison and Rowland 2005, Benner et al 2010), observer orientation (Benner et al 2010), feeding latency (Oswald and Robison 2008), and open field behavior (Oswald and Robison in review) in these strains, and shown that they vary markedly in these anxiety related behaviors.
We use a variety of genomics approaches to understand the mechanisms that drive behavioral adaptation to captivity. We have compared the brain transcriptomes of wild and domesticated strains using both microarray platforms and Illumina and 454 based RNAseq. We have discovered many promising candidate genes that appear to differentiate behaviorally bold and behaviorally shy animals. We have also used RNAseq approaches to identify more than 20,000 SNPs that vary among our zebrafish strains. We are currently using these SNPs for quantitative trait locus (QTL) analyses.
We also combine these genomics approaches with quantitative genetics experiments. Mary Oswald, a recent Ph.D. graduate from our lab, has performed selection experiments on observer orientation. She has shown that these fear related behaviors have a significant genetic basis and are genetically correlated. Her selection experiment is now being continued by Matt Singer, who is a current Ph.D. student in the lab.