Research Projects
1. Dynamin-related proteins and cytokinesis in plants
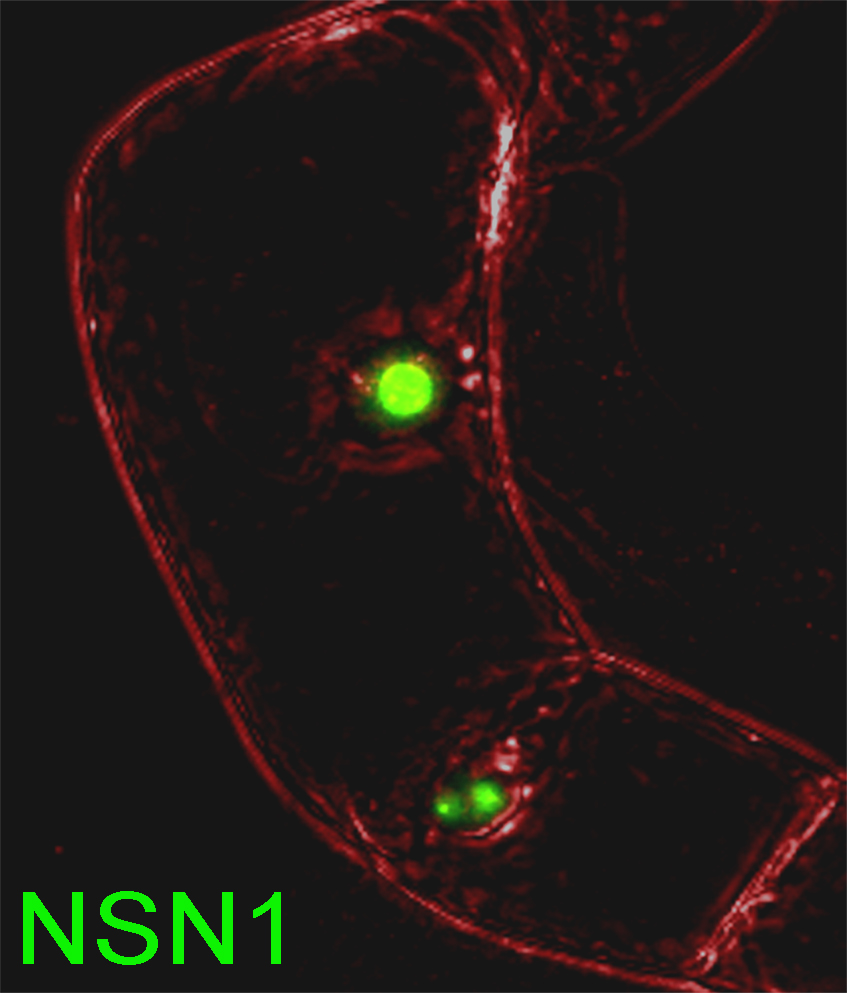
Phragmoplastin was identified from soybean as the first protein marker of the cell plate [Fig. 1] (Gu and Verma, 1995; Hong and Verma, 2008). It is a large GTPase with a molecular mass of 68 kD, and belongs to the superfamily of dynamin-related proteins (DRP) (Hong et al., 2003a). Whereas other DRP families of proteins (DRP2 through DRP6) are involved in various membrane processes in the cells [Table 1], phragmoplastins (DRP1) may function as tubulases and are required for cell plate formation and plasma membrane maintenance [Fig. 2] (Hong and Verma, 2005). The Arabidopsis genome contains five DRP1 genes, designated as DRP1A through DRP1E [Fig. 3]. Members of the DRP1 family of proteins are differentially targeted to the forming cell plate and appear to have non-redundant functions during cell plate formation [Fig. 4] (Hong et al., 2003b).
2. Callose synthesis and cell wall formation
Callose is a common name used to describe polysaccharides that show fluorescence with the aniline blue dye under UV light (Verma and Hong, 2001). Callose isolated from different types of plant cell walls may differ slightly in chemical structure and composition. In general, callose is a linear polysaccharide of several hundred glucose residues connected via beta-1,3 glycosidic bonds [Fig. 5]. Depending upon the sources of the plant material (cell wall types and plant species), callose may also contain a small percentage of glucuronic acid residues and branches via beta-1,6 glycosidic bonds. In contrast to cellulose which is beta-1,4 glucan and form crystalline structures, callose is largely helical and 'amorphous'. Structural comparable beta-1,3-glucans are present in the cell walls of yeasts, some filamentous fungi, as well as in bacteria (Stone and Clark, 1992).
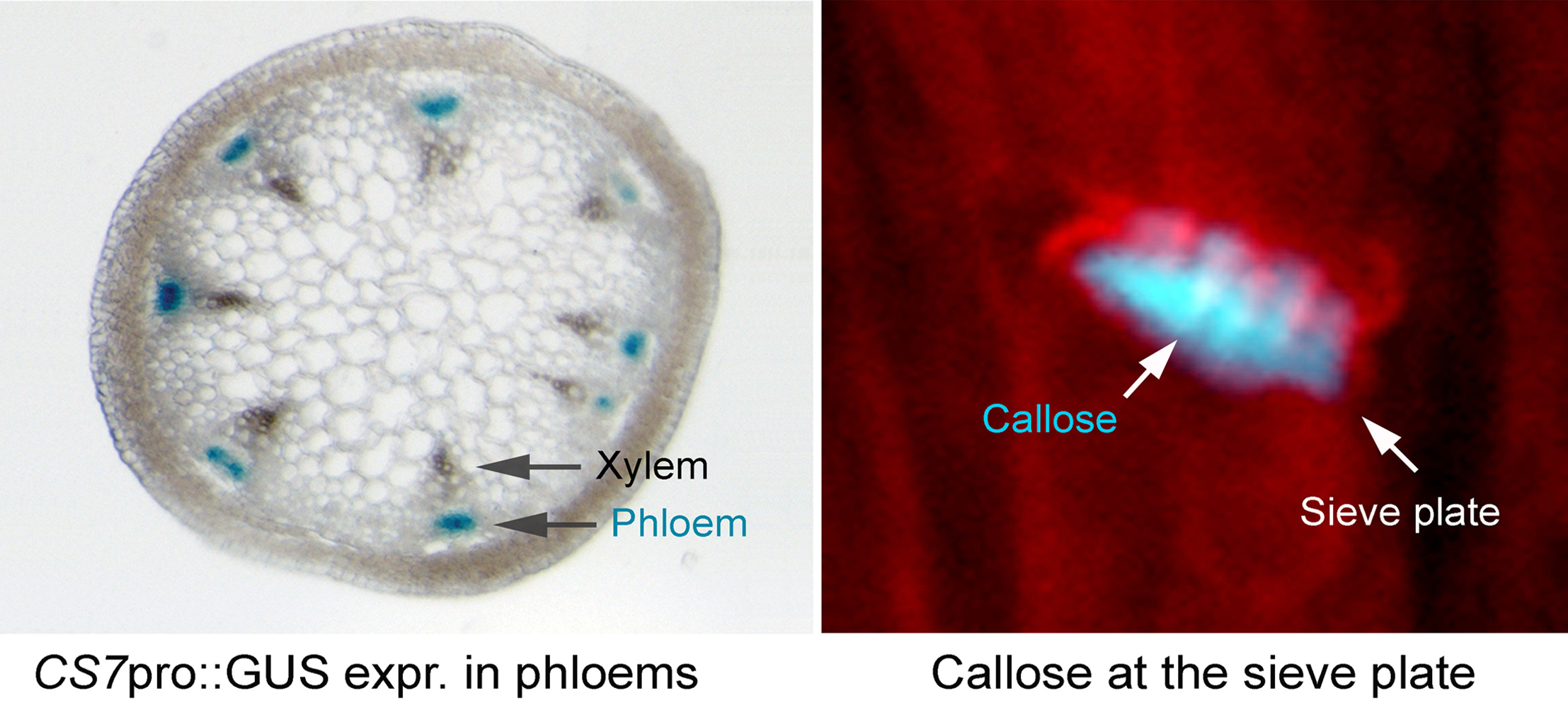
Callose is an important constituent of many specialized cell walls in plants. It is required for the formation of the cell plate, plasmodesmata, tracheids, pollen grains, pollen tubes, and root hairs. It is also produced in plant cells in response to wounding, heavy metal treatment and pathogen infection [Fig. 6] (Verma and Hong, 2001). The synthesis of this polysaccharide is catalyzed by a membrane-associated multi-subunit enzyme complex [Fig. 7]. The key component of this complex is its catalytic subunit known as callose synthase (Hong et al., 2001a). Other components of the enzyme complex may include UDP-glucose transferase (UGT1) (Hong et al., 2001b), annexin (ANN) (Andrawis et al., 1993; Shin and Brown, 1999), sucrose synthase (SuSy) (Amor et al., 1995), Rho-like small GTPase (Rop1) (Hong et al., 2001b). The list of these accessory components is growing and may vary depending upon the subcellular locations and types of callose synthases.
There are 12 callose synthase genes in the model plant Arabidopsis [Fig. 8]. They are expressed in different tissues of the plant, and respond differently to the inducers of callose synthesis, such as pathogen infection and heavy metal treatment. Callose synthase 1 (CalS1) was first identified as a cell plate-specific isoform (Hong et al., 2001a). Other ArabidopsisCalS genes are named according to their sequence homology with CalS1 [Fig. 9]. Thus, CalS2 is highly homologous to CalS1 (92%) whereas CalS12 is only distantly-related to CalS1 (35%) (Hong et al., 2001a). CalS genes have also been referred to as glucan synthase-like (GSL) (Richmond and Somerville, 2001). We prefer using the CalS nomenclature system because the gene names directly indicate their biochemical function as callose synthase catalytic subunits.
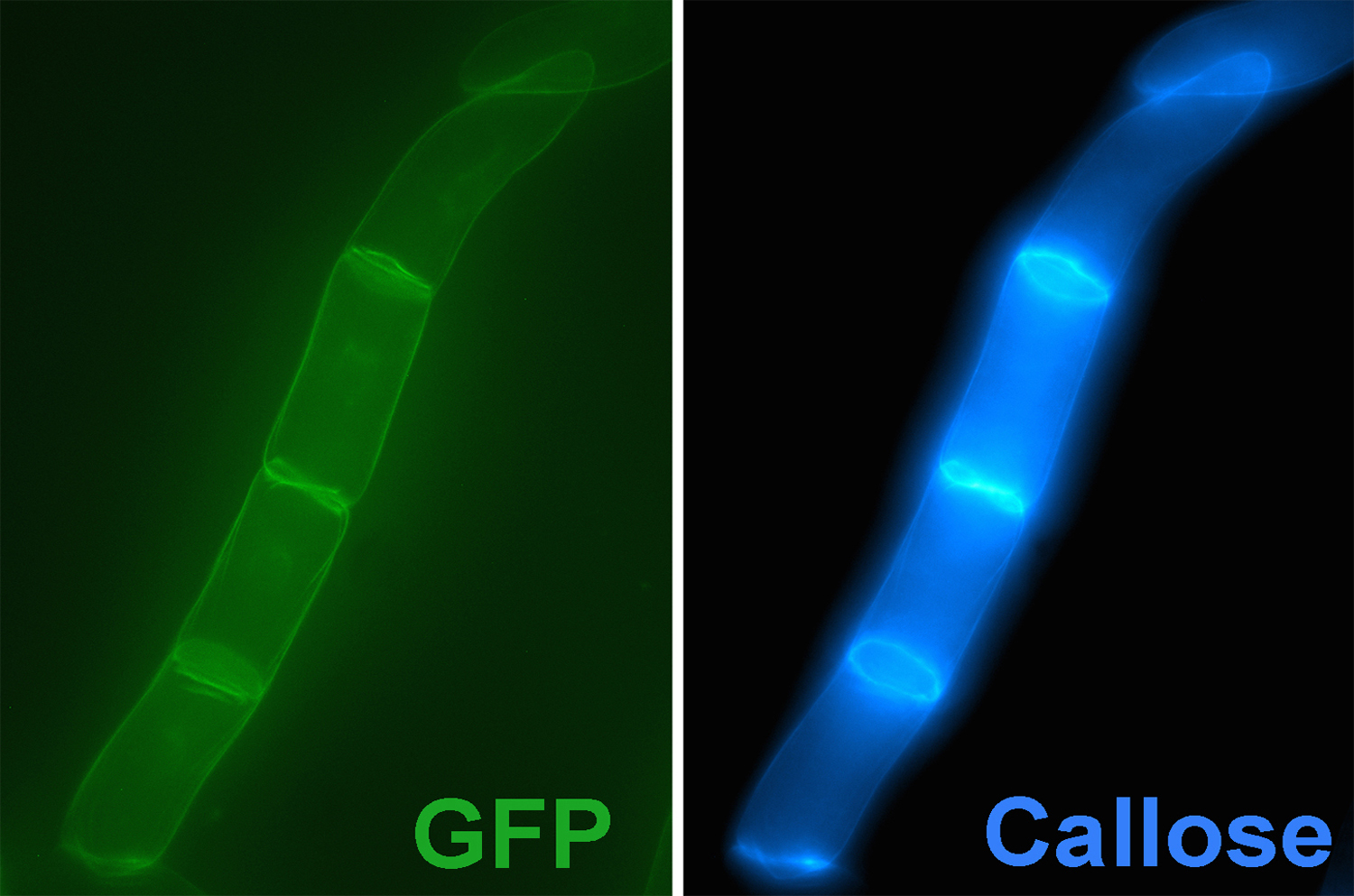
Cell plate-specific CalS1. The presence of callose at the cell plate was documented almost a half of a century ago (Eschrich, 1954). During cytokinesis in plant cells, the cell plate is formed by the fusion of vesicles derived from the Golgi apparatus. The nascent cell plate is a dynamic and fragile membranous tubular network, and is stabilized by the filling tubular structures with callose, which provides a spreading force to convert the tubular network into a plate. Cellulose (a beta-1,4-glucan) and other cell wall components are then synthesized and deposited at the expanding cell plate, leading to the formation of a new cell wall. When CalS1 is tagged with GFP, the protein accumulates in Golgi-like structures in non-dividing cells and is targeted to the forming cell plate at cytokinesis [Fig. 10]. Overexpression of CalS1 leads to the accumulation of callose at the cell plate. Interestingly, overexpression of phragmoplastin, an interacting partner of CalS1, also leads to the accumulation of callose at the cell plate, and formation of widened cell plates.
Pollen-specific CalS5. Defective pollen development and pollen tube growth were found to be associated with the T-DNA knockout mutants of cals5 [Fig. 11] (Dong et al., 2005; Nishikawa et al., 2005). No observable phenotypes could be found in the vegetative parts of cals5 mutant plants [Fig. 12]. The CalS5 gene is expressed at very high levels in developing flowers and pollen tubes. It is also expressed at relatively low levels in other parts of the plant. The roles of CalS5 in vegetative tissues are unknown. In the anthers of developing flowers, a callose wall is synthesized between the plasma membrane and the primary cell wall of the pollen mother cells. After meiosis, the microspore tetrads are encapsulated in the callose wall [Fig. 13]. Callose is also the major polysaccharide constituent of the pollen tube wall. The callose wall of pollen mother cells and microspore tetrads in cals5 mutants was very thin and lacked callose. Pollen grains were deformed and lost the characteristic surface patterning. As a consequence, over 95% of mature pollen grains were male sterile and could not germinate. Pollen tubes from the pollen grains that managed to germinate, were deformed and did not develop callose plugs. The mutant phenotypes of kompeito (kom) resemble that of the cals5 mutant. The KOM gene encodes a Rhombold-like transmembrane protease and appears to be required for the activity of CalS5 (M. Kanaoka and K. Okada, personal communication).
In addition to the cell plate-specific CalS1 and microsporogenesis-specific CalS5, the biological functions of other callose synthases have also been studied recently. CalS12 was shown to be induced upon pathogen infection (Ostergaard et al., 2002) and required for the synthesis of callose induced by bacterial infection (Nishimura et al., 2003; Jacob et al., 2003). CalS11 and CalS12 may function together during pollen development and are required for the synthesis of the callose wall that separates tetraspores (Enns et al., 2005). The functions of other CalS isoforms remain to be determined.
3. Proline metabolism, osmotolerance and stress signaling in plants
Proline is one of the 20 standard amino acids that make up proteins. It is highly water soluble and acts as a potent osmoprotectant in bacteria and higher plants. Its concentrations in plant cells vary drastically depending upon both the exogenous and endogenous physiological conditions. Proline levels in plant cells can be increased up to 400-fold upon osmotic stress [Tab. 2], and its concentrations can reach to over 200 mM in the cytoplasm of cultured tobacco cells (Binzel et al., 1987). Accumulated proline can protect enzymes and other macromolecules from denaturation, serves as an energy reserve during stress, and acts as an antioxidant against reactive oxygen species (ROS) and other free radicals.
Accumulation of proline is due primarily to de novo proline biosynthesis. In E. coli and yeast, proline is synthesized from glutamate, which is catalyzed by three enzymes, GK (gamma-glutamyl kinase), GSADH (glutamate-5-semialdehyde dehydrogenase, also known as GPR for glutamyl phosphate reductase) and P5CR (pyrroline-5-carboxylate reductase) [Fig. 14] (Csonka and Hanson, 1991). In plants, proline can be synthesized from glutamate and ornithine [Fig. 15]. The biosynthesis of proline from glutamate is catalyzed by two enzymes, P5CS (pyrroline-5-carboxylate synthetase) and P5CR (Hu et al., 1992; Delauney and Verma, 1994). The biosynthesis of proline from ornithine can occur via two routes. The first route involves the transamination of delta-NH2 group of ornithine, and is catalyzed by two enzymes, delta-OAT (ornithine delta-aminotransferase) and P5CR. The other routes involves the transamination of alpha-NH2 group, and is catalyzed by alpha-OAT (ornithine alpha-aminotransferase) and P2CR (pyrroline-2-carboxylate reductase) [Fig. 16]. Genes encoding P5CS, P5CR, and delta-OAT have been cloned (Delauney and Verma, 1994). However, genes for alpha-OAT and P2CR have yet to be cloned. Thus, proline could be synthesized from glatamate and ornithine via three pathways, i.e. the P5CS pathway, the delta-OAT1 pathway, and the alpha-OAT1 pathway [Fig. 17].
Proline levels are reduced in plant cells in response to the removal of osmotic stress. Proline is converted to glutamate, which is catalyzed by PDH (proline dehydrogenase, also known as proline oxidase) and P5CDH (pyrroline-5-carboxylate dehydrogenase) [Fig. 18]. Proline recycling can provide both C- and N-sources for the needs of rebuilding of plants during recovery from stress. Proline recycling recovers only a portion of the energy used to synthesize it.
[Reference section is under construction.]