CONTACT
Patrick J. Hrdlicka
875 Perimeter Dr MS 2343
(Renfrew Hall 307)
Moscow, Idaho 83844-2343
USA
Email: hrdlicka@uidaho.edu
Research
We are a nucleic acid chemistry group with research interests at the interface of chemistry, molecular biology, and materials science. We aim to develop molecular tools that facilitate gene regulation and detection of important diagnostic targets, and ultimately result in novel therapeutic intervention strategies against genetic diseases. Our focus is to develop:
|
|
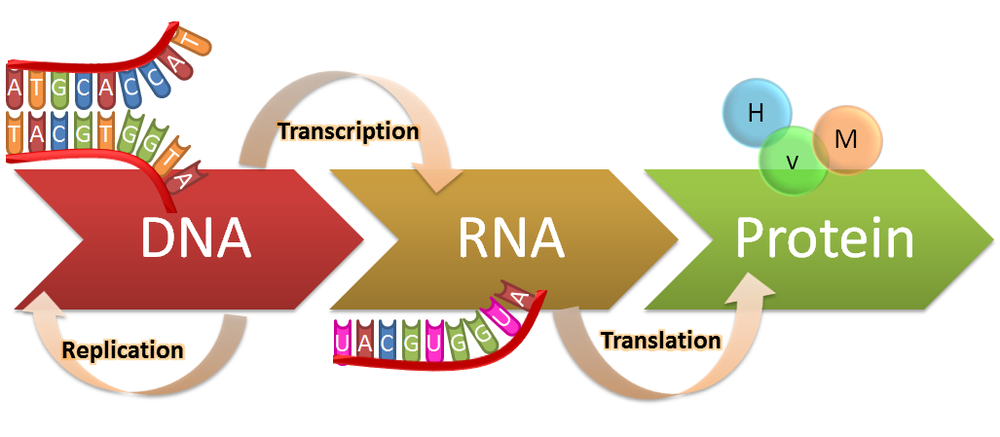
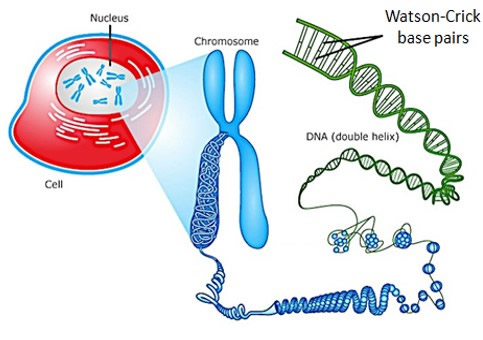
Background: Nucleic acids play a pivotal role as carriers of genetic information in all Kingdoms of Life. The genetic information of a eukaryotic cell is stored in the nucleus as double-stranded DNA. The information is processed in a highly regulated, sequence-specific, and enzyme-mediated manner to give multiple copies of a corresponding RNA transcript, which is processed and transported to the cytosol where it is used as a template for protein synthesis. Major advances over the past 15 years, have revealed that the biology of nucleic acids offers many avenues for manipulation of gene expression, which can be exploited for applications in molecular biology, biotechnology and medicine. Development of chemically functionalized oligonucleoties that enable strong, specific and stable binding to nucleic acid targets of interest is at the center of these applications.
|
DNA-recognition. The prospect of regulating gene function, detecting important biomarkers, and identifying new classes of therapeutic modalities against genetic diseases, has captivated and inspired chemists and molecular biologists to develop molecules that enable efficient recognition of specific chromosomal DNA regions. However, this has proven very challenging as the genetic information content that is contained within a specific sequence of nucleotides, is tied up in Watson-Crick base pairs and buried deeply within the compacted and nuclearly localized DNA. State-of-the-art approaches, therefore, rely on molecules that recognize sequence-specific features that are accessible via duplex groves instead. However, these strategies display limitations with respect to target sequence choice, binding specificity, and ease-of-use, which has restricted their widespread use.
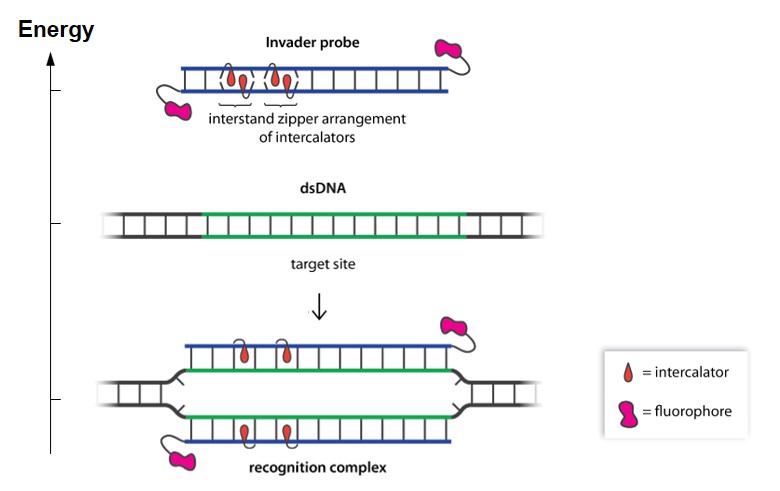
We have recently introduced a unique strategy for recognition of mixed-sequence chromosomal DNA regions, which is based on double-stranded oligonucleotide probes that are energetically activated through modification with interstrand zipper arrangements of intercalator-functionalized nucleotides. Our results suggest that these Invader probes enable efficient recognition of chromosomal DNA targets with single nucleotide specificity under non-denaturing conditions (e.g., see publication 53).
|
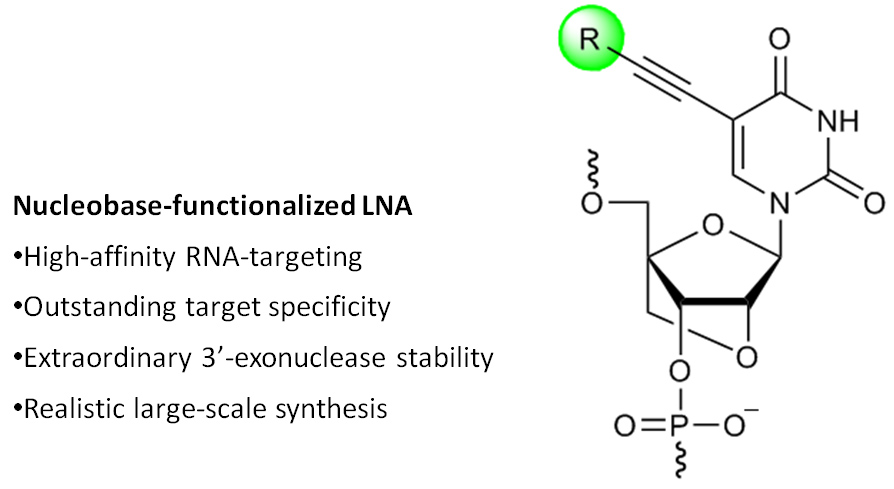
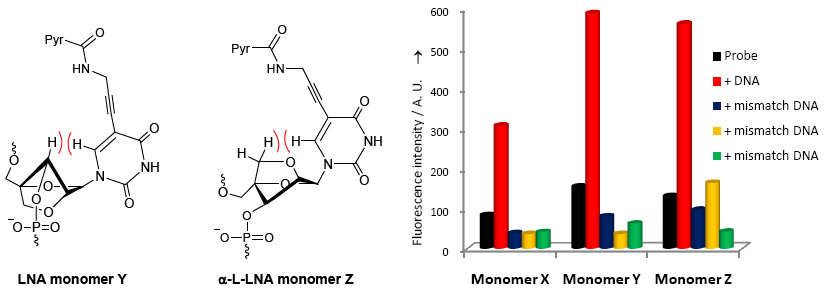
RNA-targeting oligonucleotides (e.g., antisense, siRNA, and anti-miR) are widely explored as fundamental research tools and are gaining increasing promise as therapeutic agents against diseases of genetic origin. Introduction of chemically modified nucleotides into oligos is essential to increase their binding affinity toward RNA targets, improve discrimination of mismatched RNA to reduce off-target effects, and enhance stability against nucleases to slow down degradation. Among the many chemistries that have been evaluated in the past two decades, phosphorothioate DNA (PS-DNA), O2’-alkylated RNA and affinity- and specifity-enhancing Locked Nucleic Acid (LNA) have emerged as the most therapeutically interesting modifications. The FDA approved the cholesterol-lowering antisense drug Mipomersen in January 2013. ~40 antisense drug candidates currently are evaluated in clinical trials. We are developing new nucleotide building blocks such as nucleobase-modified LNA monomers that display promising hybridization and diagnostic properties (e.g., see publication 46)
|
Nucleic acid detection probes. Reliable detection of specific nucleic acid targets is an integral component in a wide range of applications including pathogen detection, quantification of DNA/RNA in real-time PCR, visualization of RNA trafficking in cells, and detection of single nucleotide polymorphisms, which are important disease markers. The use of fluorophore-modified oligonucleotides has been a popular approach towards this end. We have developed a variety of probes and sensors that enable sensitive and specific detection of nucleic acid targets (e.g., see publication 60).
|